Iros Barozzi, PhD
Group Leader
T: +43 (0)1 40160-57517
iros.barozzi@meduniwien.ac.at
ORCID: 0000-0003-0690-3473
Research Focus
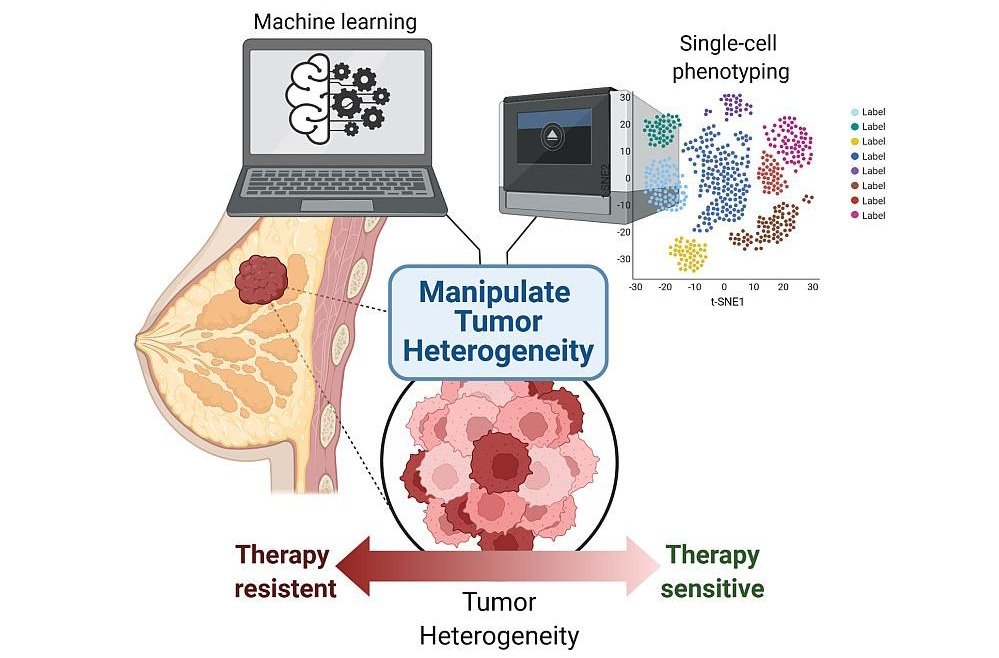
The Computational Cancer Genomics Laboratory aims at learning how gene regulatory networks are mis-regulated in the progression of breast and prostate cancers to drug-resistant, metastatic disease.
Despite difficulties in the identification of genes driving the progression to metastatic cancer, the contribution of transcriptional heterogeneity to resistance has so far received little attention compared to genetic heterogeneity.
We think that by shifting this focus we could identify and target new links between the heterogeneity of the primary tumor and resistance to therapies (Hong et al. 2019). To this aim, we are integrating information at both single-cell and population level through genomic analyses and we are using machine learning approaches to predict resistance.
These goals intercept the overarching themes of the lab, which are the identification and characterization of patterns of transcriptional heterogeneity in apparently identical cells, and the characterization of the underlying mechanisms of regulation.
We think that a better understanding of these basic mechanisms can provide a different perspective to successfully tune the treatment of primary cancer lesions, thereby preventing resistance to current therapies.
We also take advantage of our interest in computationally predicting:
- the function of non-coding, regulatory DNA (Barozzi et al. 2014, Mancino et al. 2015, Monti et al. 2017, Comoglio et al. 2019) and
- the target genes of enhancers (Gorkin et al. 2020, Osterwalder et al. 2018)
Selected Publicatons
An atlas of dynamic chromatin landscapes in mouse fetal development
Gorkin DU*, Barozzi I*, Zhao Y*, Zhang Y*, Huang H*, Lee AY, Li B, Chiou J, Wildberg A, Ding B, Zhang B, Wang M, Strattan JS, Davidson JM, Qiu Y, Afzal V, Akiyama JA, Plajzer-Frick I, Novak CS, Kato M, Garvin TH, Pham QT, Harrington AN, Mannion BJ, Lee EA, Fukuda-Yuzawa Y, He Y, Preissl S, Chee S, Han JY, Williams BA, Trout D, Amrhein H, Yang H, Cherry JM, Wang W, Gaulton K, Ecker JR, Shen Y, Dickel DE, Visel A#, Pennacchio LA#, Ren B#.
Nature. 2020 Jul;583(7818):744-751.
http://doi.org/10.1038/s41586-020-2093-3
Single-cell transcriptomics reveals multi-step adaptations to endocrine therapy
Hong SP#, Chan TE, Lombardo Y, Corleone G, Rotmensz N, Bravaccini S, Rocca A, Pruneri G, McEwen KR, Coombes RC, Barozzi I#, Magnani L#.
Nature Communications. 2019 Sep 2;10(1):3840.
http://doi.org/10.1038/s41467-019-11721-9
Dissection of acute stimulus-inducible nucleosome remodeling in mammalian cells
Comoglio F*, Simonatto M*, Polletti S, Liu X, Smale ST, Barozzi I#, Natoli G#.
Genes & Development. 2019 Sep 1;33(17-18):1159-1174.
http://doi.org/10.1101/gad.326348.119
Enhancer redundancy provides phenotypic robustness in mammalian development
Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer- Frick I, Pickle CS, Kato M, Garvin TH, Pham QT, Harrington AN, Akiyama JA, Afzal V, Lopez-Rios J, Dickel DE#, Visel A#, Pennacchio LA#.
Nature. 2018 Feb 8;554(7691):239-243.
http://doi.org/10.1038/nature25461
Limb-Enhancer Genie: An accessible resource of accurate enhancer predictions in the developing limb
Monti R*, Barozzi I*#, Osterwalder M, Lee E, Kato M, Garvin TH, Plajzer- Frick I, Pickle CS, Akiyama JA, Afzal V, Beerenwinkel N, Dickel DE, Visel A#, Pennacchio LA#.
PLoS Computational Biology. 2017 Aug 21;13(8):e1005720.
http://doi.org/10.1371/journal.pcbi.1005720
Coregulation of transcription factor binding and nucleosome occupancy through DNA features of mammalian enhancers
Barozzi I*, Simonatto M*, Bonifacio S, Yang L, Rohs R, Ghisletti S, Natoli G.
Molecular Cell. 2014 Jun 5;54(5):844-857.
http://doi.org/10.1016/j.molcel.2014.04.006
(* co-first author; # co-corresponding author)